LSD1
 LSD1_schematic
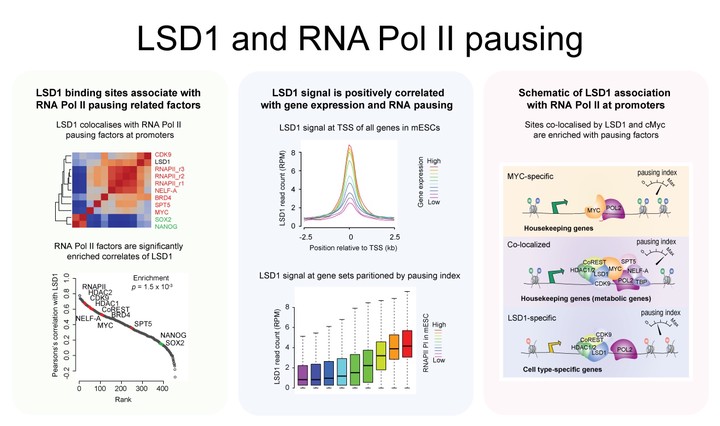
LSD1_schematicLysine-specific demethylase 1 (LSD1) plays an essential role in mammalian biology and binds to both promoters and enhancers to catalyze the demethylation at H3K4 and H3K9. In mouse embryonic stem cells (ESCs), LSD1 is well-known for its role in decommissioning enhancers during differentiation. In comparison, its role at the promoters of genes remains poorly understood in ESCs despite their widespread presence at these sites. Here, we report that LSD1 is associated with RNAPII pausing (RNAPII), a rate-limiting step in transcription regulation at the promoter-proximal region of genes, in mouse ESCs. Correlation of LSD1 binding sites against over 400 ChIP-seq datasets demonstrated a strong enrichment of factors involved in RNAP II pausing with LSD1. Previous studies have elucidated the coordinated actions of pausing and releasing factors that collectively modulate RNAPII pausing. In general, the involvement of chromatin remodellers in RNAPII pausing has not been well documented. We demonstrate that the knockdown of LSD1 preferentially affects genes with higher RNAPII pausing than those with lower pausing and, importantly, show that the co-localization of LSD1 and MYC, a factor known to regulate pause-release, is associated with the enrichment of other RNAPII pausing factors compared to their independent counterparts. Moreover, we found that genes co-occupied by LSD1 and MYC are significantly enriched for housekeeping genes that are involved in metabolic processes and globally depleted of transcription factors compared to those bound only by LSD1. These findings reveal a pleiotropic role of LSD1 in regulating housekeeping program besides its previously known role in regulating cell identity programs. Our integrative analysis presents evidence for a previously unanticipated role of LSD1 in RNAPII pausing through its association with pause release factors in modulating cell-type specific and cell-type invariant genes.